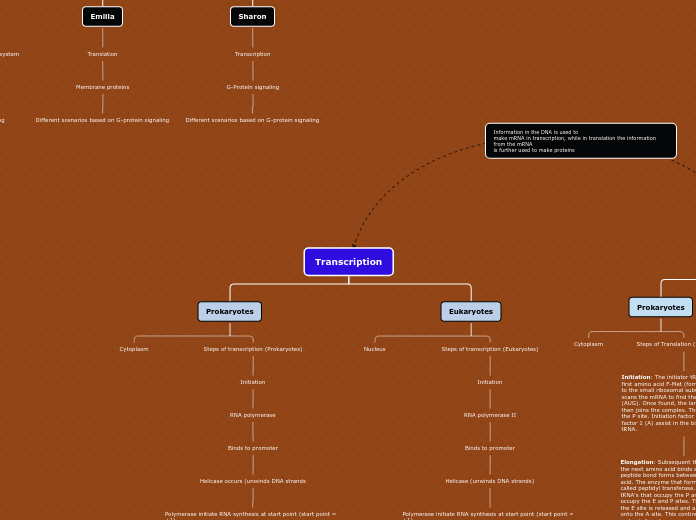
Transcription
Prokaryotes
Cytoplasm
Steps of transcription (Prokaryotes)
Initiation
RNA polymerase
Binds to promoter
Helicase occurs (unwinds DNA strands
Polymerase initiate RNA synthesis at start point (start point = +1)
RNA transcript is released and detaches from DNA
mRNA made
Eukaryotes
Nucleus
Steps of transcription (Eukaryotes)
Initiation
RNA polymerase II
Binds to promoter
Helicase (unwinds DNA strands)
Polymerase initiate RNA synthesis at start point (start point = +1)
RNA transcript is released and detaches from DNA
Pre-mRNA made
RNA processing
mRNA made
Subtopic
Translation
Prokaryotes
Cytoplasm
Steps of Translation (Prokaryotes)
Initiation: The initiator tRNA carries the first amino acid F-Met (formyl- Methionine) to the small ribosomal subunit (30s)and scans the mRNA to find the start codon (AUG). Once found, the large subunit (50s) then joins the complex. The tRNA binds to the P site. Initiation factor 3(E) and initiation factor 1 (A) assist in the binding of the tRNA.
Elongation: Subsequent tRNA which carries the next amino acid binds at the A site. A peptide bond forms between the two amino acid. The enzyme that forms the bond is called peptidyl transferase. Next, the two tRNA's that occupy the P and A sites shift to occupy the E and P sites. Then the tRNA on the E site is released and a new tRNA comes onto the A site. This continues until a stop codon is found.
Termination: Once the stop codon is reached, a release factor arrives onto the A site and helps separate the complex.
Eukaryotes
Cytoplasm
Steps of Translation (Eukaryotes)
Initiation: The mRNA for eukaryotes differs in that it contains factors such as CAP on the 5' end and a poly A tail on the 3' end. This occurs only in eukaryotes because eukaryotes travel a longer distance from the nucleus to the cytoplasm for translation while transcription and translation for prokaryotes both occur in the cytoplasm.
The initiator tRNA carries the first amino acid Met (Methionine) to the small ribosomal subunit (40s)and scans the mRNA to find the start codon (AUG). Once found, the large subunit (60s) then joins the complex. The tRNA binds to the P site.
Elongation: Subsequent tRNA which carries the next amino acid binds at the A site. A peptide bond forms between the two amino acid. The enzyme that forms the bond is called peptidyl transferase. Next, the two tRNA's that occupy the P and A sites shift to occupy the E and P sites. Then the tRNA on the E site is released and a new tRNA comes onto the A site. This continues until a stop codon is found
Termination: Once the stop codon is reached, a release factor arrives onto the A site and helps separate the complex.
Eukaryotic Cells
Organelles: Structure and Function
Mitochondria: powers and produces energy for the cell.
Cytoplasm: jelly-like, keeps everything in place, and protects cell organelles from getting damage.
Microtubules: move vesicles and organelles throughout the cell.
Lysosome: digests waste, and cleans up the cell
Smooth ER: makes lipids and steroids.
Centrosome: organizes microtubules
Chromatin: proteins and DNA
Nucleolus:creates ribosomes, is located around the nucleus
Nuclear Envelope: protein lined; allows movement in and out the nucleus
Ribosomes:help synthesizeprotein, carry genetic info, and helps repair damages.
Nucleus:conttrol center of the cell, houses DNA
Peroxisomes: metabolizes waste.
Rough ER: makes membrane proteins, waste, and has ribosomes.
Microfilaments: proteins (fibrous) that form the cellular cortex.
Plasma Membrane: protects, structures and detects transport in and out the cell.
Golgi apparatus: modifies proteins.
Vacuole: like a trash can, collects waste.
Pathway of how proteins enters the endomembrane system
The amino acids helps determine the proteins destination. Proteins with the ER signal gets escorted into the Rough ER with the help of SPR (signal recognition particle).
Organelles
Cytoplasm
Carries nutrients
Helps receive chemical signals
Translates chemical signals into intracellular functions
Mitochondria
Proteins break down sugars, and help produce cellular energy.
Perioxisomomes
Newly synthesized proteins are focused on degradation of lipids on the peroxisome surface.
Chloroplasts (plants)
Nucleus
Involved in transcription factors, RNA polymerase, DNA polymerase, and DNA binding proteins
Plastids (plants)
Carry transit peptides.
Rough ER
Golgi Apparatus makes lysosomes
Lysosomes carry proteins out using exocytosis
List of Membrane Proteins
Membrane transport protein: translocates solutes, ions, nutrients, neurotransmitters, and drugs.
Translocase: Changes ATP for ADP in the intermembrans space.
Potassium Channel: Allows K+ to diffuse freely across the membrane, but not other ions, such as Na+ or Cl-
Ligand gated ion channel: allow for specified ions to pass through the membrane. (Like: Na+, K+, Ca2+, Cl-)
Cytochrome: An iron-containing protein that is a component of electron transport chains in the mitochondria and chloroplasts of eukaryotic cells and the plasma membranes of prokaryotic cells
ATP Synthase:
produces ATP
Glycoprotein:
initiate immune responses
Integrin:
links the cytoskeleton
to the extracellular matrix
Aquaporin:
transports water
across cell membranes
Receptor Tyrosine Kinase:
involved in cellular communication
G Protein Signaling
Reception
Signaling molecule binds to the receptor
Transduction
Relay molecule in signal transduction pathway
Response
Activation of cellular response
Different scenarios based on G protein signaling:
Phosphorylation Cascade:
Signaling molecule binds to Receptor
Relay molecule activates protein kinase 1
Kinase takes a phosphate group and inactivates protein kinase 1
Active Protein kinase 1 activates protein kinase 2 by converting ATP to ADP
Kinase removes a phosphate group from active protein kinase and results in a active protein kinase 2
Active protein kinase 2 phosphorylates a protein that brings about the cell's response to the signal
Active Protein kinase 2 is converted from a ATP to a ADP - active protein is formed
Protein phosphatases catalyze the removal of the phosphate groups from the proteins, making the proteins inactive again
The completion of this cycle results in a cellular response
Cellular response = Gene expression = mRNA = make protein
cAMP as a second messenger in a G protein signaling pathway:
Signaling molecule binds to GPCR
G Protein is activated (GDP goes to GTP)
Activated G protein goes into the adenylyl cyclase
cAMP activates another protein and leads to a cellular response
Contributions:
Victoria:
Pathway of how proteins enter the endomembrane system
Eukaryotic Cell
Different scenarios based on G-protein signaling
Emilia
Translation
Membrane proteins
Different scenarios based on G-protein signaling
Sharon
Transcription
G-Protein signaling
Different scenarios based on G-protein signaling
Different types of responses can come from a signal molecules. When activated, the responses assist in the synthesis of a certain protein. Multiples responses can result in more of a particular gene being expressed.
Information in the DNA is used to
make mRNA in transcription, while in translation the information from the mRNA
is further used to make proteins