Nucleoid
Capsules
Fimbrae
Pili
Endospore
Plasmid
Cell Wall
Slime Layers
Glycoacalyx
In common with
Eukaryotes
Ribosomes
Cytoplasm
This hots the cellular respiration that happens in map two in order to produce energy.
Flagellum
Plasma Membrane
Lipids
ex: cholesterol
Essential component in
PLASMA MEMBRANE
Made of phospholipds
Carbohdrates
ex: glucose
Nucleic Acids
DNA+RNA
Proteins:
ex: actin
Protein structures
Protein has 4 levels of structure
Primary
peptide bonds
Amino end bonds
to carboxyl end
Secondary
hydrogen bonds
alpha and beta sheets
Tertiary
ionic and covalent bonds; disulfide bonds
interaction of r groups
Quaternary
ionic and covalent bonds; disulfide
R-groups
polar
phospholipid
unsaturated
liquid at room temp
double bonds
ester linkage
fatty acid
cis
kinks
hydrogenation
trans
saturated
solid at room temp
no double bonds
cysteine
acidic
glutamic
basic
histidine
non polar
glycine
FOUND IN PLANTS
Cell Wall
Chloroplasts
Plasmodesmata
FOUND IN PLANTS,
ANIMALS, AND
FUNGI
Cytoskeleton
Cell Membrane
Nucleus
Nuclear Envelope
Nucleolus
Endoplasmic Reticulum
Smooth
Rough
Mitochondria
Peroxisomes
Vacuoles
In common with
Prokaryotes
Ribosomes
Cytoplasm
Flagellum
Plasma Membrane
FOUND IN ANIMALS
Lysosomes
Centrsome
Cilia
Plants
photosynthesis
Mesophyll interior tissue of leaf
Chloroplast roughly 30-40 within each mesophyll
Light Reactions serves to convert solar energy to chemical energy, receiving products from Calvin Cycle
Thylakoid Membrane, is where this process occurs,
Photosystem II p680,contains more chlorophyll b, noncyclic phosphorilation
Cyclic Electron Flow- cycles electrons through PSI n n nm n, , m Non Cyclic Electron Flow- occurs in both PSI and PSII and electrons received from water^
Photosystem I p700, contains more chlorophyll a, involved in cyclic phosphorylation
Calvin Cycle serves to synthesize sugar made from products received from Light Reactions
Stroma, is where this process occurs and is found in the inner space of the chloroplast
Phase II: Reduction during this phase the products from phase I: Carbon fixation, 3 PGA, as well as 6 ATP and 6 NADPH →Out puts: 6 ADP, 6 NADP+ and 6 G3P, 1 is used to make Glucose, 5 move on to next step and are recycl^
Phase III: Regeneration uses 5 G3P, 3 ATP and coverts that to regenerate the RuBP acceptor creating 3 RuBp and 3 ADP and thus cycle repeats, phase I
Animals
Glycolysis
(cytosol)
Process: Breakdown sugar, and has energy investment phase and energy payoff phase
Glucose + 2 ATP + 2 NAD+ = 2 Pyruvate,
2 NADH, 4 ATP
NET= 2 PYRUVATE, 2 NADH, 2 ATP
Step One: Hexokinase converts Glucose
to G6P
Step Three: Phosphofructokinase Fructose 6 Phosphate converted to Fructose 1, 6 Bisphosphate
ATP Production: Substrate Level Phosphorylation
Creates the ATP needed by DNA Polymerase
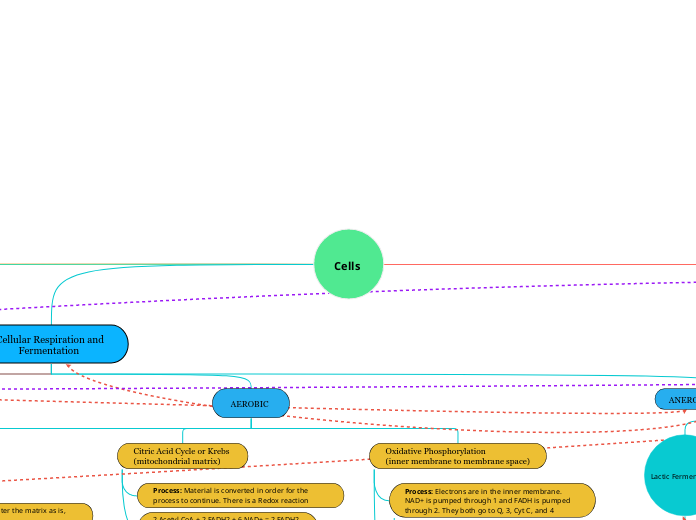
AEROBIC
Pyruvate Oxidation
(cytosol to matrix)
Process: Pyruvates cannot enter the matrix as is, they must convert
2 Pyruvate + 2 CoA + 2 NAD+ = 2 Acetyl CoA &
2 NADH
NET= 2 ACETYL COA & 2 NADH
Important: The cycle is ran twice because pyruvate is not produced singularly
ATP Production: There is none
Citric Acid Cycle or Krebs
(mitochondrial matrix)
Process: Material is converted in order for the process to continue. There is a Redox reaction
2 Acetyl CoA + 2 FADH2 + 6 NAD+ = 2 FADH2,
6 NADH, 2 ATP
NET: 2 FADH2, 6 NADH, 2 ATP
Step One: Acetyl CoA + Oxaloacetate makes Citrate
Step Three: Isocitrate makes Alpha Ketoglutarate (redox reaction)
ATP Production: Substrate Level Phosphorylation
Oxidative Phosphorylation
(inner membrane to membrane space)
Process: Electrons are in the inner membrane. NAD+ is pumped through 1 and FADH is pumped through 2. They both go to Q, 3, Cyt C, and 4
The energy decreases as it goes on. Small amounts of energy are constantly released to stop lysing.
Protons rush beside it and their associated energy creates powers the creation of ATP
O2 + 10 NADH+ 2 FADH2 = H2O + 26-28 ATP
NET: H20 + 26-28 ATP
Key Step: ATP Synthase from ATP with an endergonic reaction. ADP + Pi
ATP Production: Chemiosmosis and Electron Transport Chain coupling
ANEROBIC (FERMENTATION)
Lactic Fermentation
lactate
Alcohol Fermentation
ethanol
NADH-> NAD+
Membrane Receptors
receptors are embedded
into the plasma membrane
G Protein
Reception
A signal molecule attaches to
the extracellular side
of a G protein coupled receptor,
causing GCPR to change shape
Cytoplasmic side of GCPR
binds to a G protein
G protein is now activated
and carries a GTP molecule
Activated G protein diffuses along
plasma membrane and binds to
an enzyme, adenylyl cyclase,
activating it
G protein phosphatase function:
removes a phosphate from GTP,
reverting it back to GDP
GDP: G protein is INACTIVE
GTP: G protein is ACTIVE
Transduction
Adenylyl cyclase synthesizes
cyclic AMP from ATP
(2 phosphate groups are lost)
cAMP is the second messenger,
or a relay molecule
cAMP activates previously inactive
protein kinase 1
Protein kinase 1 activates
protein kinase 2
(by addition of phosphate)
ATP is converted to ADP
Protein phosphatases
inactivate protein kinase 1
Protein kinase 2 activates
protein kinase 3 and so on
Phosphodiesterase
converts cAMP to AMP
Response
The last protein kinase in
the phosphorylation cascade
brings about response in cell
Response is AMPLIFIED
one signal molecule produces
more and more molecules
at each step of cascade
Response occurs in
cytoplasm or nucleus
Last kinase enters nucleus and
activates transcription factor,
transcripts gene into mRNA
mRNA directs protein synthesis
Tyrosine Kinase
Signal molecules bind to two
separate polypeptides, inactive
tyrosine kinase proteins
2 polypeptides DIMERIZE:
come together
AUTOPHOSPHORYLATION:
polypeptides function as kinases,
they each take phosphate groups
from ATP and add them to it's partner
(phosphate is added to tyrosines)
ACTIVATED tyrosine kinase receptors
are ready to interact with relay proteins
Intracellular Receptors
receptors are inside the cell
Signal molecule easily passes
through nonpolar (hydrophobic)
membrane
Signal binds to receptor (a protein) in
cytoplasm
Signal bound receptor enters the nucleus
and binds to specific genes
Receptor protein acts as a transcription factor
and transcripts gene into mRNA
mRNA is translated into a specific protein
Eukaryotes
Located in the nucleus
Numerous amount of ORI
linear DNA
Replication is bidrectional and discontinuous
Origin of replication- two strands of DNA at ORI sequences and form a bubble
Visual
ORI
Leading stand
Forks out 5'
Lagging strand
Forks out 3'
Subtopic
Enzymes involved
Toperisomerase: helps relieve any strain caused by unwinding of the DNA
Helicase:untwists the double helix of DNA at the replication forks.
Single Stranded Binding Proteins: keeps DNA single stranded
Primase: joins RNA nucleotides to make the primer.
Prokaryotes
Located in the Cytoplasm
Only one replication bubble
Circular DNA
Replication is bidirectional
DNA Polymerase I and III
enzymes used
also involved
Occurs in the cytoplasm
Proteins are made from code in mRNA
Enzyme aminoacyl-tRNA synthetase cataylyzes
the bonding of an amino acid to a specific tRNA
Initiation
The small ribosomal subunit
binds to mRNA (first the 5' cap and then scans mRNA to find the start codon)
An initiator tRNA bound to the small ribosomal subunit with the correct anticodon base pairs with the mRNA. This tRNA carries the amino acid methionine, MET in eukaryotes and formylmethionine, FMET in prokaryotes
The large ribosome subunit, with E, P, and A sites then binds to the small, forming the translation initiation complex. Initiation factors are proteins needed for this process.
The initiator tRNA is now in the P site.
Elongation
The anticodon of an incoming
tRNA into the A site binds with the complimentary
mRNA codon.
A peptide bond forms with the help of the enzyme
peptidyl transferase between the new amino acid attached to the tRNA in the A site and the C end of the polypeptide in the P site. The polypeptide is removed from the tRNA in the P site and transferred to the tRNA in the A site.
The tRNA in the A site moves to the P site and the empty tRNA in the P site moves to the E site, where it exits. The mRNA is moving along allowing for the next codons to be exposed so that anticodons can bind in the A site.
mRNA is read from 5' to 3' and amino acids are joined together in the N to C direction (Amino end to carboxyl end).
Termination
When a stop codon is reached in the A site,
there is no tRNA to match this codon.
A release factor sits in the A site
and stops translation.
Location is signaled based on the codons
Secretory Pathway
The use of signal molecules is similar to the use of signaling in map 2 done by the G-Protein.^
Polypeptide creation on a free ribosome
SRP stops synthesis for a while by binding to the peptide
SRP binds to an Endoplasmic Reticulum Receptor
SRP leaves which allows for synthesis to continue and some of the protein is inside of the ER
The signal is removed by an enzyme and the protein is left in the cell
Plants
Can go immediately to the mitochondria, nucleus, peroxisomes, and chloroplast
Order of Pathway
Nucleus- Creation
ER- leaves shipped through a vesicle
Golgi Bodies- package up proteins in order for them to be secreted.
Removed using spliceosomes because they do not code for anything
Joined together once introns are removed
Glycolosis
Leading Strand
Forks out 5'
Complementary DNA strand synthesized continuously 5' to 3' direction
DNA polymerase III, synthesis this strand
Sliding Clamp, changes DNA POL III from being distributive (falling off) to processive (staying on)
RNA primers, is what the strand is composed of
Okazaki gragments are what extend this RNA primer
DNA Polymerase I will remove RNA primers with new nucleotides (DNA)
Ligase enzyme: with phosphodiester bonds will make seam less bond