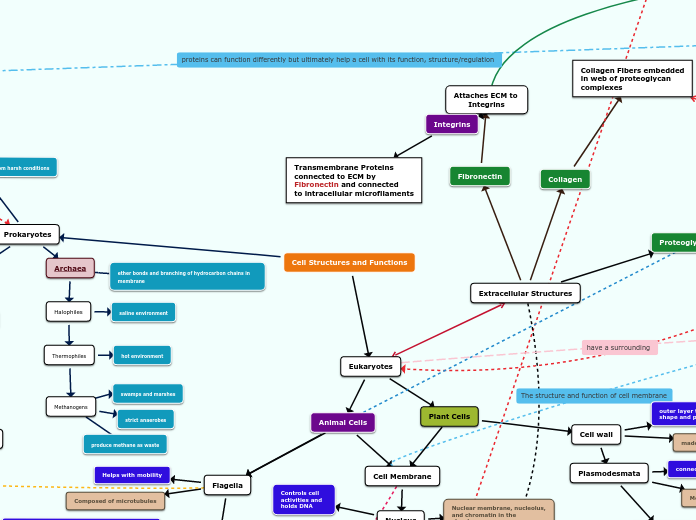
Extracellular Structures
Fibronectin
Attaches ECM to
Integrins
Collagen
Collagen Fibers embedded
in web of proteoglycan
complexes
Proteoglycan
Forms Proteoglycan Complex
Animal Cells
Cell Membrane
Nucleus
Nuclear membrane, nucleolus, and chromatin in the structure
Controls cell activities and holds DNA
Mitochondria
Has an inner and outer membrane, and also a matrix.
Generates ATP and where cellular respiration takes place
Smooth E.R.
Tubular membrane like structure; lacks ribosomes
Synthesizes lipids, detoxifies, metabolizes carbohydrates, and stores calcium
Rough E.R.
membrane continuous with nuclear membrane and has ribosomes attached to it
Makes proteins and transports vesicles
Ribosomes
Made of RNA and protein
Synthesizes proteins
Peroxisome
Made of a membrane and proteins
produced hydrogen peroxide as a by product and then coverts it to water
Golgi Apparatus
Made of cisternae (flattened sacked pouches)
packages and processes lipids and proteins; receives and releases cargo
Cytoskeleton
made up of microtubules, actin filaments, and intermediate filaments (all made of proteins)
holds the cell together, helps the cell to keep its shape, and aids in movement
Vacuole
Thin membrane with fluid inside
In animal cells they are small and used for waste. In plant cells they are large and help maintain water balance and food
Flagella
Helps with mobility
Composed of microtubules
Centrosome
Region where cell's microtubules are initiated
Composed of two centrioles (the protein tubulin)
Lysosome
Hydrolyze Macromolecules
Has a membrane and is a sphere
Microvilli
Projections that increase cell's surface area
Covered in plasma membrane
Plant Cells
Cell wall
outer layer that maintains cell's shape and protects cell
made of cellulose, other polysaccharides, and protein
Plasmodesmata
connect the cytoplasms of adjacent cells
Membrane line channels
Chloroplast
Converts energy of sunlight to chemical energy
Inner and outer membrane, and a granum made of thylakoid
Archaea
ether bonds and branching of hydrocarbon chains in membrane
Halophiles
saline environment
Thermophiles
hot environment
Methanogens
swamps and marshes
strict anaerobes
produce methane as waste
Bacteria
Plasma membrane
Nucleoid
Cell wall
Capsules and slime layers
Flagella
Fimbriae
Pili
connection; transfer of DNA
short and help the cell attach to surfaces
movement
protect the cell from ingestion and digestion of white blood cells
contains peptidoglycan
contains genetic material of the cell
encloses the organelles of the cell
can form endospores for protection from harsh conditions
no nucleus, no membrane-bound organelles
Extracellular Matrix Ecm
Phospholipid bilayer
hydrophilic head w/ 2 hydrophobic fatty acid tails
Polar head w/ phosphate group
Membrane fluidity
Saturated
Hydrocarbons tails VISCOUS (gel-like) & tighly packed together
Unsaturated
Hydrocarbon tails w kinks (FLUID) & not tightly packed together
Affected by Temperature
Increase/HIgh Temp.
Decrease/Low Temp.
Cholestrol
Helps maintain membrane fluidity & more VISCOUS
Carbohydrates
Stick out from membrane to attach to proteins & define cell's characteristics
Help cells identify chemical signals
Proteins
functions
Enzyme Activity
Signal Transduction
Cell-Cell Recognition
Intracellular Joining
Transport
Attach to ECM & Cytoskeleton
Integral/Transmembrane
Integral: a-helix w/ R-group protrude out from backbone
Peripheral
on surfcae of membrane b/c hydrophilic R-group
Smaller # of molecules
Passive Transport
No energy to move from high to low down conc'n gradient
Simple Diffusion
Osmosis (hypertonic, hypotonic, isotonic)
Facilitated Diffusion
Chanel & Carrier Proteins
transport hydrophilic/polar/charged sustances
Active Transport
Uses energy to move from low to high against conc'n gradient
examples
Na+/K+ Pump
2 K+ ions go inside where K+ levels are high & pumps out 3 Na+ out
uneven charge distribution creates voltage across membrane
Proton Pump
generates proton gradient across memebrane
Contransport
active transport of solute indirectly drives transport of other substances
Bulk transport
pinocytosis
A cell takes fluid inside the cell
phagocytosis
Food particles gather at the surface of the cell
larger # of molecules
Endocytosis
Exocytosis
- Removes waste - Intake nutrients - Releases synthesized molecules - Neurotransmitters receptors - Hormonal triggers
Hydrophobic/nonploar molecules cross easily
Ribose sugar
One-Stranded
Contains Uracil (A-U)
Acts as messenger between DNA & ribosomes to make amino acids & proteins
Deoxyribose sugar
Double-Stranded
Conatins Thymine (A-T)
Stores & transfers genetic info
Nitrogenous bases, deoxyribose sugar, phosphate group
Hydrogen bonds between complemetary base pairs
Phosphodiester bond between pentose sugar (-OH group) and phosphate group
Nucleotides
Transcription
1) Initiation
1. RNAP binds to promoter.
DNA strands unwind.
RNAP- Enzyme
RNA Polymerase
2.With DNA unwound, RNA synthesis starts at start point (+1) on template strand.
Start point can be
on promoter!
2) Elongation
1. RNAP moves downstream
unwinding DNA and elongating
RNA from 5' to 3' direction.
RNAP decodes the DNA and creates the RNA transcript complement from the template strand.
2. DNA strands reform
double helix on their own
as RNAP moves downstream.
3) Termination
1. Eventually RNA transcript released.
RNAP detaches from DNA.
Translation
Enzyme aminoacyl-tRNA synthase attaches correct amino acid to corresponding tRNA 3' end
tRNA-decodes specific codons of mRNA using anticodon in order for correct amino acid to be added
Occurs in cytoplasm
don't have introns and happens in cytoplasm
Transcription
1. Initiation
RNA polymerase II binds to the promoter by transcription factors. The RNA polymerase then separates the DNA into single strands so that the template can be read in the 3' -> 5' direction.
promoter is also called TATA box
2. Elongation
Pre-mRNA nucleotides are paired with their complementary bases which correspond with the template strand of DNA. The pre-mRNA moves in the 5' -> 3' direction and the template DNA strand moves in the 3' -> 5' direction.
pre-mRNA has a 5' cap
this is where small subunits of ribosome attach and scan
pre-mRNA has a poly-A tail
keeps RNA secure by adding 100-200 A's
pre-mRNA does not contain thymine and instead contains uracil which is used for the complementary base for adenine
3. Termination
When the RNA polymerase II reaches the terminator it stops and detaches from the DNA
this is where RNA processing or splicing occurs : introns are removed and all exons are formed together to form the final mRNA.
the spliceosome removes the introns and joins together the exons
final (mature) mRNA moves from nucleus to cytoplasm
once they are separated the two DNA strands come back together to reform the double helix
the new mRNA molecule leaves the nucleus and attaches to a ribosome
uses ATP
Translation
1. Initiation
Small ribosomal subunit binds to an mRNA
Initiator tRNA binds to the start codon on the mRNA with its anti-codon base pairs
Start codon is AUG
tRNA binds to the P site first
tRNA carries Methionine so it can find the start codon
The large ribosomal subunit arrives and binds to the small subunit, tRNA, and mRNA to form the initiation complex.
Hydrolysis of GTP provides energy for this process
2. Elongation
The next tRNA binds to the A site and a peptide bond forms between the two amino acids.
Peptidyl Transferase forms the bonds between the two amino groups
The tRNA is now moved from the P site to the E site to be removed and then the tRNA that was at the A site is moved to the P site. This happens over and over bringing more tRNA's to the A site with a chain of amino acids attached to it.
The mRNA is read in 5' to 3' direction and amino acids are added from N terminal to C terminal.
3. Termination
Once the stop codon is reached in the A site a release factor sits in the A site and stops translation
Binds to the 5' cap
The enzyme Amino acyl tRNA synthetase will pair the tRNA and amino acid correctly
Main components of Translation are Ribosomes and tRNA
Ribosomes have three binding sites for tRNA. They are the P site (Peptidyl-tRNA), E site (exit site), and A site (Aminoacyl-tRNA binding site)
Not present in Prokaryotes
happens in the nucleus and has introns
polypeptide synthesis begins on the free ribosome in the cytosol
A SRP (signal recognition protein) binds to the signal peptide which briefly stops synthesis
The SRP binds to a receptor protein in the ER.
The SRP detaches from the receptor, in which protein synthesis starts up again (translocation across membrane)
The signal peptide is split by an enzyme in the receptor protein complex called signal peptidase
the final polypeptide leaves the ribosome and folds into its' final form
other destinations in which proteins can leave from are: mitochondria, chloroplasts, peroxisomes, nucleus (in organelles) and can also leave from the cytoplasm
Proteins in the membrane can function to:
Facilitate Diffusion
Maintain Concentration
Gradients
Work in Active Transport
Work in Cell to Cell
Communication
Help Activate Cell
Signaling Pathways
path taken by a protein in a cell on synthesis to modification and then released out of cell (secretion)
3. Termination
1. no tRNA that corresponds to stop codon once it reaches A-site
2. Release factors sits at A-site which then causes the complex to break apart
Gene Organization
Operon
A cluster of functionally related genes that are in the same pathway and can be regulated together by an operator
An operator is usually within the promoter and allows proteins (Activators and Repressors) to turn gene expression on and off (like a light switch)
When a repressor is bound to the operator gene expression is off and is the regular basal level.
When an activator is bound to the operator gene expression is on and at a high level
Promoter is a sequence of DNA that is needed to turn gene expression on and off and controls how RNA polymerase binds to it
Operator
Lac Operon
The lac operon is organized with Structural genes, Regulatory gene, and Regulatory regions
The regulatory gene is the lac I gene
The structural genes are the lac Z, lac Y, and lac A genes
The proteins permease and B-galactosidase are expressed from lac Y and lac Z and are used to make glucose and galactose
lac Z, lac Y, and lac A genes are induced by the presence of lactose
The regulatory region are the promoter and operator
E. coli can grow with glucose and lactose and it uses the lac operon to do so.
Glucose present
Lactose Present
constitutive
the genes in the lac operon encode proteins that allow for bacteria to use lactose as energy
Gene Regulation
when there is no lactose available
the repressor is made and binds to the operator
the lac operon is off (basal)
example of negative regulation
when there is only lactose present
the repressor binds to lactose
adenylyl cyclase is active
CAP is active
lac operon is on
expression is at high level
RNA polymerase binds to promoter for transcription for lac operon genes
lactose present but no glucose
repressor bound to lactose
adenylyl cyclase is active
CAP is active
cAMP levels high
lac operon is on
when on, all structural genes are transcribed to form a long mRNA
the mRNA forms 3 proteins: B-galactosidase, permase, and transacetylase
B-galactosidase breaks down the glycosidic linkage to form glucose and galactose
CAP helps RNAP to bind promoter to facilitate transcription
when there is only glucose present
the lac repressor is bound to the operator
adenylyl cyclase is inactive
CAP is inactive
lac operon is off
both glucose and lactose present
lac repressor is bound to lactose
adenylyl cyclase is inactive
CAP is inactive
lac operon is off
cAMP levels are low
cannot help RNAP bind to the promoter
Gene Organization
Gene Regulation
Transcription Factors bind to DNA promoter region (TATA box). RNA poly II binds & unwinds DNA double helix to begin transcription
Transcription Factors
General/Basal TFs
Protein that binds promoter region & express background/low levels of transcription
binds to Proximal Control Elements
sequence upstream of gene close to promoter
Specific TFs
Activators
Positive control over gene expression as they increase levels of transcription
Origin of these
Specific TF's are from
Cell Signaling Pathways!
These are activated as the last
molecule in Transduction phase of
the Signaling Pathway
Repressors
30nm Fiber
10nm Fiber
DNA winds around Histones
forming Nucleosome Beads.
Strung together by Linker DNA
Nucleosome Interactions
cause thin fiber to coil into
thicker fiber
Forms Looped Domains that
Attach to Proteins
Step 1)
Step 2)
Step 3)
Fully Activated Dimer complex can now
interact with relay proteins to trigger a
CELLULAR RESPONSE!
Transduction
Once Tyrosine Kinases are Dimerized,
they autophosphorylate each other by
taking a phosphate off of ATP and add
it to the tyrosine on the receptor.
Dimer Complex Fully Activated!
Overall Components
Two Receptor Tyrosine-Kinases
Tyrosine Attached to
Receptors
Start off as Monomers.
Become Dimers once both
activated.
Kinases: Can add phosphate groups
to each other. A process called
Autophosphorylation.
Signal Molecules
ATP
Relay Proteins
first the signal molecule binds to the GPCR
this causes the receptor to modify its shape and activate in order to bind to the g protein
the activated g protein coupled receptor binds to the g protein
this causes the bound GDP to convert into GTP which activates the g protein
the activated g protein and GTP binds to the adenylyl cyclase
the GTP is hydrolyzed (water is added to it) and it activates the adenylyl cyclase
the activated adenylyl cyclase converts ATP to cAMP
cAMP activates another protein (could be a protein kinase) which leads to multiple cellular responses
cAMP is the second messenger
once a relay molecule activates a protein kinase it starts a signal transduction pathway (phosphorylation cascade)
a relay protein activates a protein kinase 1 which activates a protein kinase 2
the active protein kinase 2 phosphoryates an inactive/active protein that brings about the cell's repsonse to the signal
protein phosphates catalyze the removal of phosphate groups from proteins making the proteins inactive again
this happens again and again resulting in the cascade
This results in an amplification of the effect of the signal molecule.
ATP is turning into ADP
ATP comes from the mitochondria and other cellular processes like Substrate Level Phosphorylation and Oxidative Phosphorylation
signal molecule is the first messenger
reception stage
Reception